Chapter 28 Filtered human PBMCs (10X Genomics)
28.1 Introduction
This performs an analysis of the public PBMC ID dataset generated by 10X Genomics (Zheng et al. 2017), starting from the filtered count matrix.
28.2 Data loading
28.3 Quality control
Cell calling implicitly serves as a QC step to remove libraries with low total counts and number of detected genes. Thus, we will only filter on the mitochondrial proportion.
library(scater)
stats <- high.mito <- list()
for (n in names(all.sce)) {
current <- all.sce[[n]]
is.mito <- grep("MT", rowData(current)$Symbol_TENx)
stats[[n]] <- perCellQCMetrics(current, subsets=list(Mito=is.mito))
high.mito[[n]] <- isOutlier(stats[[n]]$subsets_Mito_percent, type="higher")
all.sce[[n]] <- current[,!high.mito[[n]]]
}qcplots <- list()
for (n in names(all.sce)) {
current <- unfiltered[[n]]
colData(current) <- cbind(colData(current), stats[[n]])
current$discard <- high.mito[[n]]
qcplots[[n]] <- plotColData(current, x="sum", y="subsets_Mito_percent",
colour_by="discard") + scale_x_log10()
}
do.call(gridExtra::grid.arrange, c(qcplots, ncol=3))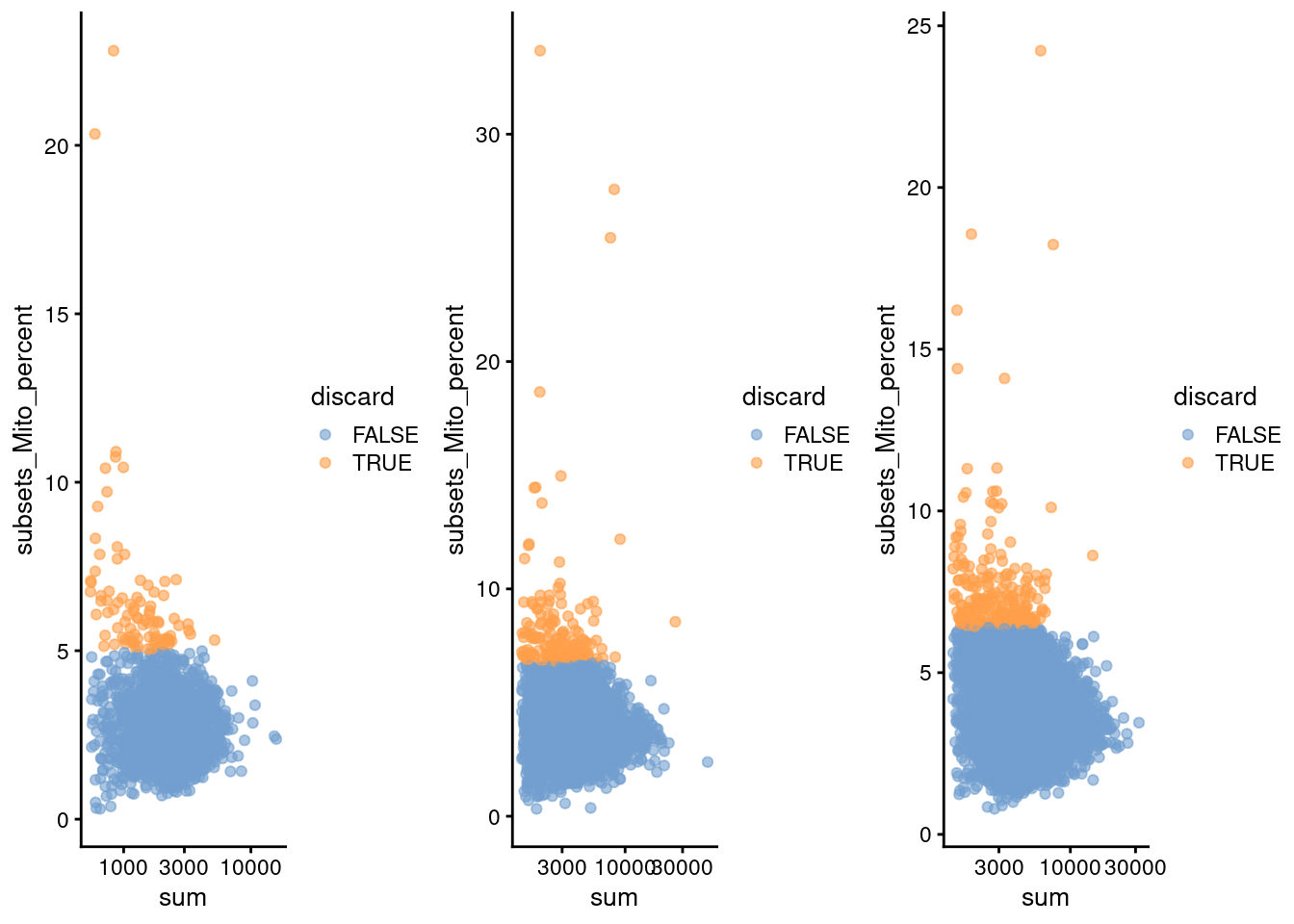
Figure 28.1: Percentage of mitochondrial reads in each cell in each of the 10X PBMC datasets, compared to the total count. Each point represents a cell and is colored according to whether that cell was discarded.
## $pbmc3k
## Mode FALSE TRUE
## logical 2609 91
##
## $pbmc4k
## Mode FALSE TRUE
## logical 4182 158
##
## $pbmc8k
## Mode FALSE TRUE
## logical 8157 22428.4 Normalization
We perform library size normalization, simply for convenience when dealing with file-backed matrices.
## $pbmc3k
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.234 0.748 0.926 1.000 1.157 6.604
##
## $pbmc4k
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.315 0.711 0.890 1.000 1.127 11.027
##
## $pbmc8k
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.296 0.704 0.877 1.000 1.118 6.79428.5 Variance modelling
library(scran)
all.dec <- lapply(all.sce, modelGeneVar)
all.hvgs <- lapply(all.dec, getTopHVGs, prop=0.1)par(mfrow=c(1,3))
for (n in names(all.dec)) {
curdec <- all.dec[[n]]
plot(curdec$mean, curdec$total, pch=16, cex=0.5, main=n,
xlab="Mean of log-expression", ylab="Variance of log-expression")
curfit <- metadata(curdec)
curve(curfit$trend(x), col='dodgerblue', add=TRUE, lwd=2)
}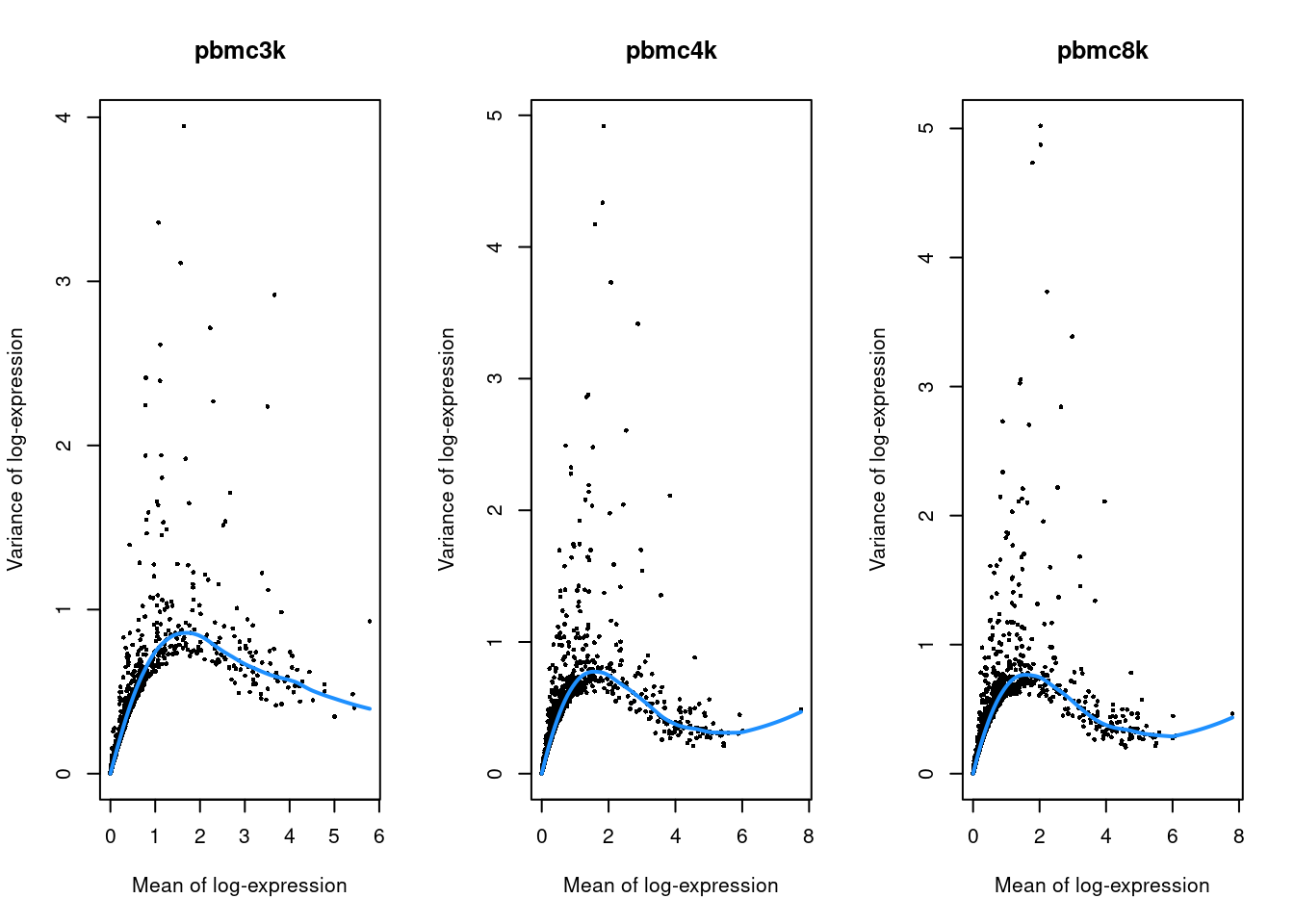
Figure 28.2: Per-gene variance as a function of the mean for the log-expression values in each PBMC dataset. Each point represents a gene (black) with the mean-variance trend (blue) fitted to the variances.
28.6 Dimensionality reduction
For various reasons, we will first analyze each PBMC dataset separately rather than merging them together. We use randomized SVD, which is more efficient for file-backed matrices.
library(BiocSingular)
set.seed(10000)
all.sce <- mapply(FUN=runPCA, x=all.sce, subset_row=all.hvgs,
MoreArgs=list(ncomponents=25, BSPARAM=RandomParam()),
SIMPLIFY=FALSE)
set.seed(100000)
all.sce <- lapply(all.sce, runTSNE, dimred="PCA")
set.seed(1000000)
all.sce <- lapply(all.sce, runUMAP, dimred="PCA")28.7 Clustering
for (n in names(all.sce)) {
g <- buildSNNGraph(all.sce[[n]], k=10, use.dimred='PCA')
clust <- igraph::cluster_walktrap(g)$membership
colLabels(all.sce[[n]]) <- factor(clust)
}## $pbmc3k
##
## 1 2 3 4 5 6 7 8 9 10
## 487 154 603 514 31 150 179 333 147 11
##
## $pbmc4k
##
## 1 2 3 4 5 6 7 8 9 10 11 12 13
## 497 185 569 786 373 232 44 1023 77 218 88 54 36
##
## $pbmc8k
##
## 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
## 1004 759 1073 1543 367 150 201 2067 59 154 244 67 76 285 20 15
## 17 18
## 64 9all.tsne <- list()
for (n in names(all.sce)) {
all.tsne[[n]] <- plotTSNE(all.sce[[n]], colour_by="label") + ggtitle(n)
}
do.call(gridExtra::grid.arrange, c(all.tsne, list(ncol=2)))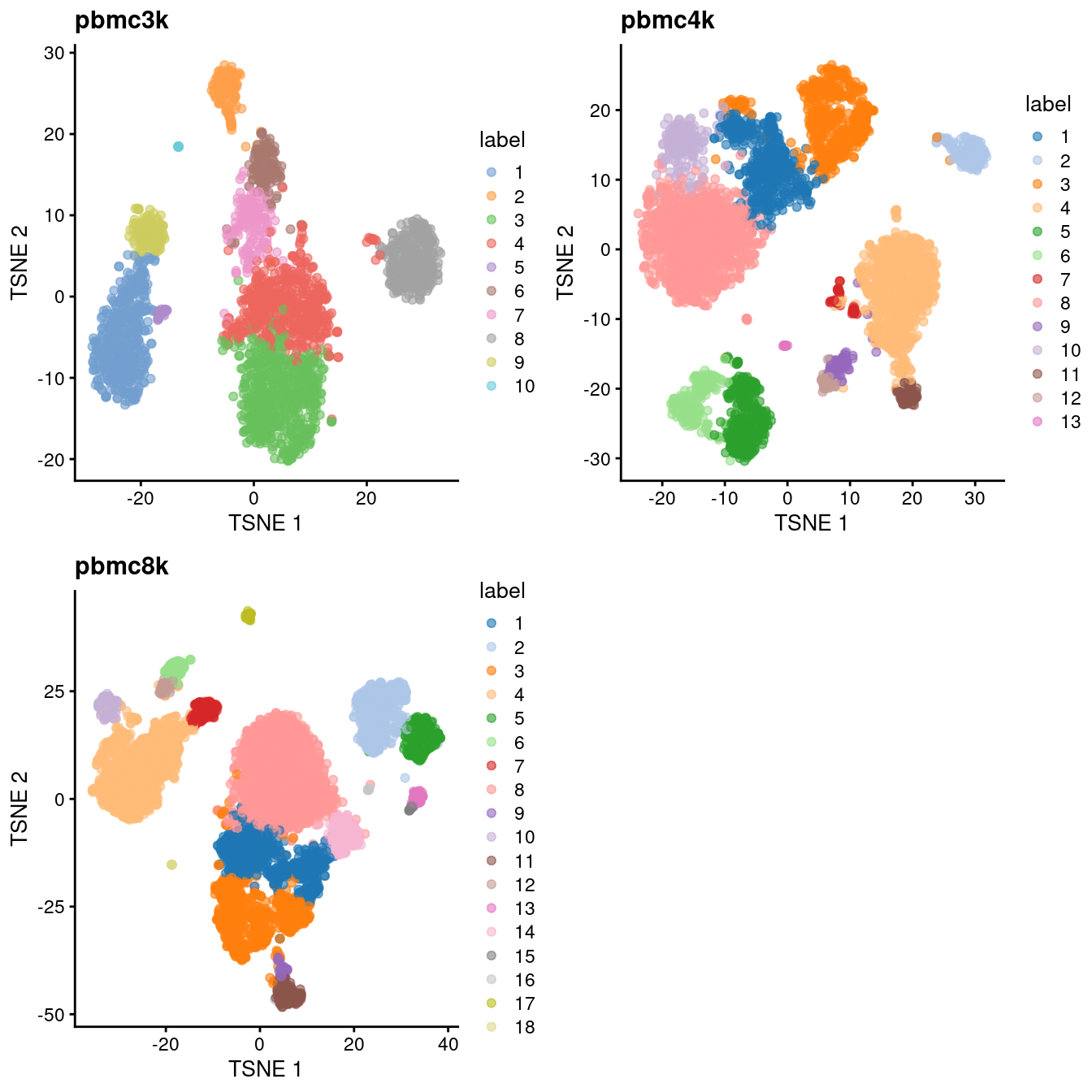
Figure 28.3: Obligatory \(t\)-SNE plots of each PBMC dataset, where each point represents a cell in the corresponding dataset and is colored according to the assigned cluster.
28.8 Data integration
With the per-dataset analyses out of the way, we will now repeat the analysis after merging together the three batches.
# Intersecting the common genes.
universe <- Reduce(intersect, lapply(all.sce, rownames))
all.sce2 <- lapply(all.sce, "[", i=universe,)
all.dec2 <- lapply(all.dec, "[", i=universe,)
# Renormalizing to adjust for differences in depth.
library(batchelor)
normed.sce <- do.call(multiBatchNorm, all.sce2)
# Identifying a set of HVGs using stats from all batches.
combined.dec <- do.call(combineVar, all.dec2)
combined.hvg <- getTopHVGs(combined.dec, n=5000)
set.seed(1000101)
merged.pbmc <- do.call(fastMNN, c(normed.sce,
list(subset.row=combined.hvg, BSPARAM=RandomParam())))We use the percentage of lost variance as a diagnostic measure.
## pbmc3k pbmc4k pbmc8k
## [1,] 7.003e-03 3.126e-03 0.000000
## [2,] 7.137e-05 5.125e-05 0.003003We proceed to clustering:
g <- buildSNNGraph(merged.pbmc, use.dimred="corrected")
colLabels(merged.pbmc) <- factor(igraph::cluster_louvain(g)$membership)
table(colLabels(merged.pbmc), merged.pbmc$batch)##
## pbmc3k pbmc4k pbmc8k
## 1 113 387 825
## 2 507 395 806
## 3 175 344 581
## 4 295 539 1018
## 5 346 638 1210
## 6 11 3 9
## 7 17 27 111
## 8 33 113 185
## 9 423 754 1546
## 10 4 36 67
## 11 197 124 221
## 12 150 180 293
## 13 327 588 1125
## 14 11 54 160And visualization:
set.seed(10101010)
merged.pbmc <- runTSNE(merged.pbmc, dimred="corrected")
gridExtra::grid.arrange(
plotTSNE(merged.pbmc, colour_by="label", text_by="label", text_colour="red"),
plotTSNE(merged.pbmc, colour_by="batch")
)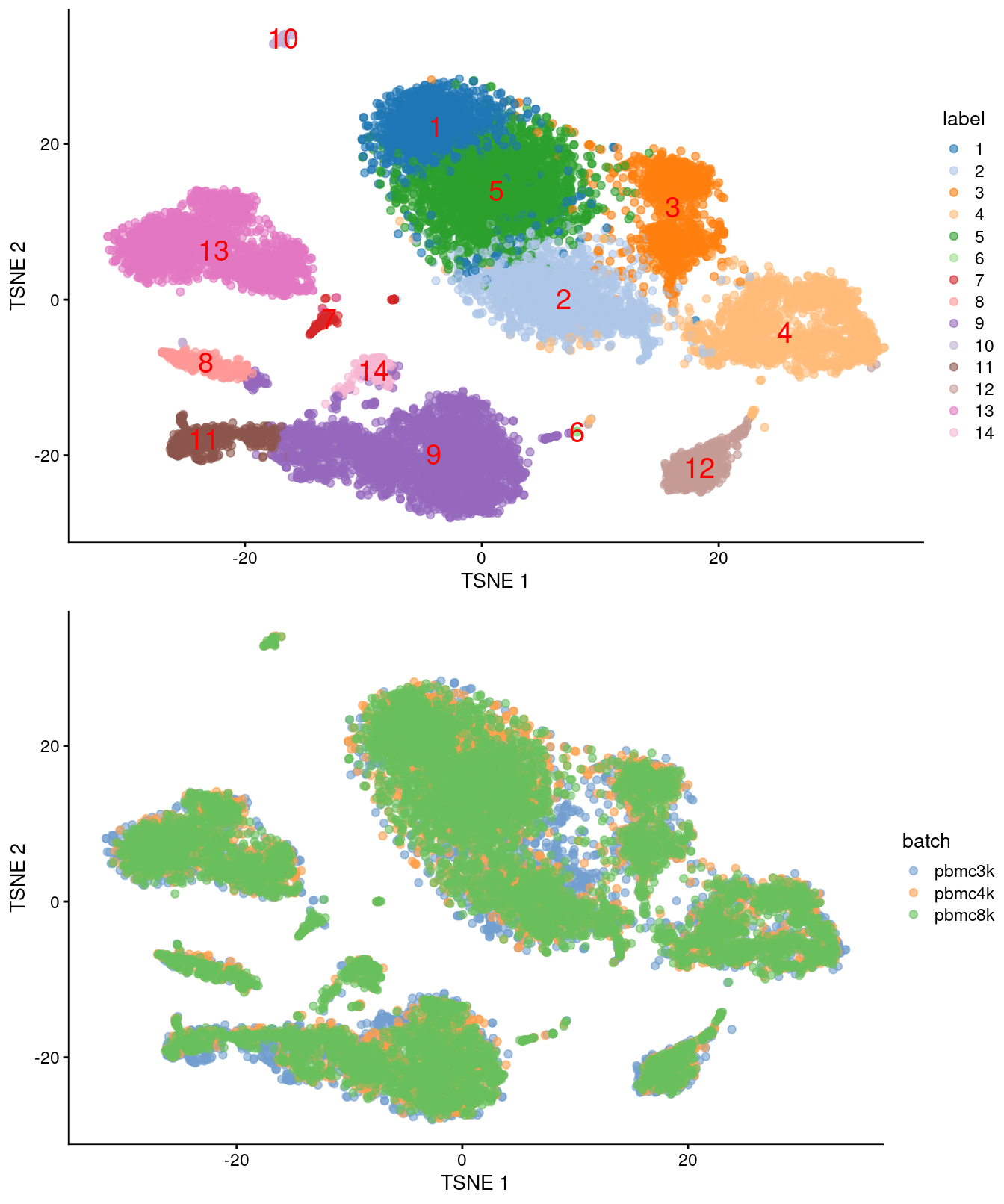
Figure 28.4: Obligatory \(t\)-SNE plots for the merged PBMC datasets, where each point represents a cell and is colored by cluster (top) or batch (bottom).
Session Info
R version 4.0.4 (2021-02-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.2 LTS
Matrix products: default
BLAS: /home/biocbuild/bbs-3.12-books/R/lib/libRblas.so
LAPACK: /home/biocbuild/bbs-3.12-books/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=C
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] batchelor_1.6.2 BiocSingular_1.6.0
[3] scran_1.18.5 scater_1.18.6
[5] ggplot2_3.3.3 TENxPBMCData_1.8.0
[7] HDF5Array_1.18.1 rhdf5_2.34.0
[9] DelayedArray_0.16.2 Matrix_1.3-2
[11] SingleCellExperiment_1.12.0 SummarizedExperiment_1.20.0
[13] Biobase_2.50.0 GenomicRanges_1.42.0
[15] GenomeInfoDb_1.26.4 IRanges_2.24.1
[17] S4Vectors_0.28.1 BiocGenerics_0.36.0
[19] MatrixGenerics_1.2.1 matrixStats_0.58.0
[21] BiocStyle_2.18.1 rebook_1.0.0
loaded via a namespace (and not attached):
[1] AnnotationHub_2.22.0 BiocFileCache_1.14.0
[3] igraph_1.2.6 BiocParallel_1.24.1
[5] digest_0.6.27 htmltools_0.5.1.1
[7] viridis_0.5.1 fansi_0.4.2
[9] magrittr_2.0.1 memoise_2.0.0
[11] limma_3.46.0 colorspace_2.0-0
[13] blob_1.2.1 rappdirs_0.3.3
[15] xfun_0.22 dplyr_1.0.5
[17] callr_3.5.1 crayon_1.4.1
[19] RCurl_1.98-1.3 jsonlite_1.7.2
[21] graph_1.68.0 glue_1.4.2
[23] gtable_0.3.0 zlibbioc_1.36.0
[25] XVector_0.30.0 Rhdf5lib_1.12.1
[27] scales_1.1.1 DBI_1.1.1
[29] edgeR_3.32.1 Rcpp_1.0.6
[31] viridisLite_0.3.0 xtable_1.8-4
[33] dqrng_0.2.1 bit_4.0.4
[35] rsvd_1.0.3 ResidualMatrix_1.0.0
[37] httr_1.4.2 FNN_1.1.3
[39] ellipsis_0.3.1 pkgconfig_2.0.3
[41] XML_3.99-0.6 farver_2.1.0
[43] scuttle_1.0.4 CodeDepends_0.6.5
[45] sass_0.3.1 uwot_0.1.10
[47] dbplyr_2.1.0 locfit_1.5-9.4
[49] utf8_1.2.1 tidyselect_1.1.0
[51] labeling_0.4.2 rlang_0.4.10
[53] later_1.1.0.1 AnnotationDbi_1.52.0
[55] munsell_0.5.0 BiocVersion_3.12.0
[57] tools_4.0.4 cachem_1.0.4
[59] generics_0.1.0 RSQLite_2.2.4
[61] ExperimentHub_1.16.0 evaluate_0.14
[63] stringr_1.4.0 fastmap_1.1.0
[65] yaml_2.2.1 processx_3.4.5
[67] knitr_1.31 bit64_4.0.5
[69] purrr_0.3.4 sparseMatrixStats_1.2.1
[71] mime_0.10 compiler_4.0.4
[73] beeswarm_0.3.1 curl_4.3
[75] interactiveDisplayBase_1.28.0 tibble_3.1.0
[77] statmod_1.4.35 bslib_0.2.4
[79] stringi_1.5.3 highr_0.8
[81] ps_1.6.0 RSpectra_0.16-0
[83] lattice_0.20-41 bluster_1.0.0
[85] vctrs_0.3.6 pillar_1.5.1
[87] lifecycle_1.0.0 rhdf5filters_1.2.0
[89] BiocManager_1.30.10 jquerylib_0.1.3
[91] RcppAnnoy_0.0.18 BiocNeighbors_1.8.2
[93] cowplot_1.1.1 bitops_1.0-6
[95] irlba_2.3.3 httpuv_1.5.5
[97] R6_2.5.0 bookdown_0.21
[99] promises_1.2.0.1 gridExtra_2.3
[101] vipor_0.4.5 codetools_0.2-18
[103] assertthat_0.2.1 withr_2.4.1
[105] GenomeInfoDbData_1.2.4 grid_4.0.4
[107] beachmat_2.6.4 rmarkdown_2.7
[109] DelayedMatrixStats_1.12.3 Rtsne_0.15
[111] shiny_1.6.0 ggbeeswarm_0.6.0 Bibliography
Zheng, G. X., J. M. Terry, P. Belgrader, P. Ryvkin, Z. W. Bent, R. Wilson, S. B. Ziraldo, et al. 2017. “Massively parallel digital transcriptional profiling of single cells.” Nat Commun 8 (January): 14049.